Immunoassays to measure SARS-CoV-2 antibodies in serum (Part II)
Serology assays that detect antibodies to SARS-CoV-2 in human serum are critical to delivering data that provides insight into immune responses, measures health impact, and supports vaccines and therapeutics R&D. Important factors to consider are the assay targets and the best platform to ensure reliable data can be generated efficiently and with a minimum of resources, including time and reagents.
The search for the most informative epitopes
Serology assays are needed to follow the infection in the population and individuals. Possible antigen reagents for serology tests include the spike (S), envelope (E), membrane (M) and nucleocapsid (N) structural proteins of the SARS-CoV-2 virus (Figure 1). The S and N proteins have been employed in a number of serology assays. One specific region that has been focused on is the receptor binding domain (RBD) of the S protein, which is directly involved in SARS-CoV-2 binding to the human angiotensin-converting enzyme 2 (ACE2) receptor for entry into host cells and is an active target for neutralizing antibodies and vaccines. The SARS-CoV-2 nucleocapsid protein (N-protein) antibody may often only indicate exposure to the virus, and not provide protection against reinfection.
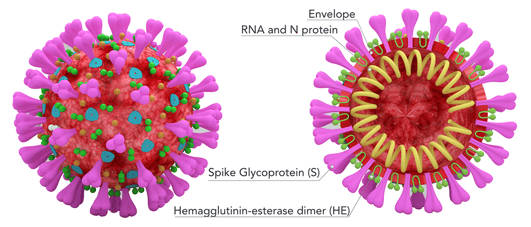
Figure 1 – Diagram of SARS-CoV-2 virus particle with possible antigen structures labeled
The receptor binding domain (RBD) of the S protein has become a key target for analysis. The RBD of the spike protein in SARS-CoV-2 is poorly conserved between SARS-CoVs and other pathogenic coronaviruses, which means that antibodies against this domain are likely to be highly specific to SARS-CoV-2 (1). There is also a strong correlation between levels of RBD-binding antibodies and SARS-CoV-2 neutralizing antibodies in patients (1).
Research carried out at the University of Texas MD Anderson Cancer Center, USA has given us more insight into the value of measuring antibodies against SARS-CoV-2 S-RBD (2). A serological enzyme-linked immunosorbent assay (ELISA) using SARS-CoV-2 S-RBD and N-protein was used to determine the levels of circulating antibodies in serum samples from confirmed COVID-19 hospitalized patients and also healthy and non-COVID-19 serum samples that were collected between June 2017 and June 2020. Their results showed that 1.6% of healthy and non-COVID-19 samples collected during the pandemic were positive for the S-RBD antibody, of which 86% had neutralizing capacity. In contrast, while 3% had the N-protein antibody, only 74% of these samples had neutralizing capacity. A majority (96.5%) of the individuals that were positive for both antibodies exhibited neutralizing capacity.
"These findings suggest that the presence of the S-RBD antibody is the best indicator of any potential protection against reinfection," said senior author Raghu Kalluri, M.D., Ph.D., professor and chair of Cancer Biology in an interview with Science Daily (3). The researchers believe that accurate and reliable S-RBD serological testing is needed to identify individuals with neutralizing antibodies and drive the development of methods to fight the pandemic.
Seroconversion indicates the need to determine both IgM and IgG
Seroconversion in COVID-19 does not necessarily follow the IgM to IgG path common to viral diseases. A review of recent pre-prints indicates that IgM and IgG levels increase from 3 to 14–28 days post illness onset (Figure 2), and there is no set chronology to seroconversion (4). This has also been observed in patients infected with SARS-CoV and Middle East Respiratory Syndrome coronavirus (MERS-CoV). These results highlight the need to measure both IgM and IgG antibodies to confirm an infection.
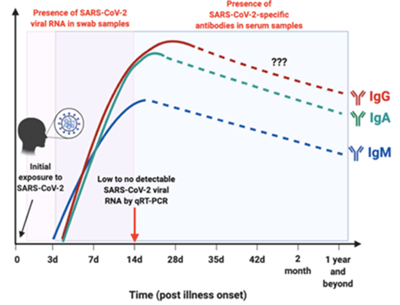
Figure 2. The profiles for viral RNA or antibodies detected in SARS-CoV-2-infected individuals. Redrawn from Figure 2, Lee et al, 2020 (reference 4)
Assay performance is critical
One of the great challenges in all serology testing is developing assays with high specificity and sensitivity, which has resulted in a lot of discussion about the reliability and utility of serological antibody tests used for COVID-19. A particular problem is cross-reactivity of antibodies between coronaviruses, for example between SARS-COV-2 and one of the many coronaviruses that cause common colds. The result is false positives. Even a test with 100% sensitivity and 99% specificity, a true antibody prevalence of 1% would result in one half of all positive results being false positives (5). Similarly, with infection prevalence at 10% and sensitivity and specificity at 90%, still only half of positive results would be true positives.
The challenge of rapid generation of high-quality data based on low reagent consumption
There is therefore a need for an assay to determine total antibodies generated against the spike protein, especially the RBD of SARS-CoV-2 in human serum samples. The question is, what is the best platform to use?
The need to quickly generate data has led to the development of a wide range of methods based on enzyme-linked immunosorbent assay (ELISA), chemiluminescence enzyme immunoassays (CLIA), fluorescence immunoassays (FIA), and lateral flow immunoassays (LFIA). A systematic review and meta-analysis that evaluated IgM and IgG tests based on ELISA, CLIA, FIA, and LFIA indicated that tests using the S antigen appear to be more sensitive than tests based on the N antigen (6). Tests that measure IgG perform better compared to those measuring IgM, while combined IgG/IgM tests seem to provide higher sensitivity than measuring either antibody alone. All the methods tested deliver high specificity with some of them (ELISA and LFIA) reaching levels around 99%. The sensitivity of methods based on ELISA and CLIA was in the range 90–94%, compared with 80–89% for LFIA and FIA.
Finding a more effective alternative to ELISA serological tests
While the immunoassay workhorse ELISA can certainly deliver reliable data, there are issues with the high consumption of reagents and samples, plus the limitations of lengthy and laborious manual workflows that take many hours to deliver data. There is therefore the need for open platforms that enable data to be rapidly generated using small amounts of reagent and sample, preferably over a broad analytical range to minimize the need for repeat analysis. To meet this need, Gyros Protein Technologies has developed a Gyrolab® immunoassay for the qualitative detection of total antibodies generated against the receptor binding domain (RBD) of the spike (S) protein of SARS-CoV-2 in human serum samples. This is the subject of the final article in this series.
Find out more by downloading the White Paper: One-hour, microfluidic SARS-CoV-2 antibody immunoassay using Sino Biological reagents and Gyrolab® immunoassay platform
References:
- The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Premkumar, L et al. Sci. Immunol.10.1126/sciimmunol.abc8413 (2020).
- Heterogeneous antibodies against SARS-CoV-2 spike receptor binding domain and nucleocapsid with implications for COVID-19 immunity. McAndrews, KM et al. JCI Insight. 2020;5(18):e142386. https://doi.org/10.1172/jci.insight.142386.
- Frequently used serology test may not detect antibodies that could confirm protection against reinfection of COVID-19, study shows. Science Daily. August 14, 2020. https://www.sciencedaily.com/releases/2020/08/200814163313.htm
- Serological approaches for COVID-19: Epidemiologic perspective on surveillance and control. Lee CY-P, et al. Front. Immunol. 2020. 11:879.doi: 10.3389/fimmu.2020.00879
- COVID-19: Understanding the science of antibody testing and lessons from the HIV epidemic. Weiss SH & Wormser, GP. Diagnostic Microbiology and Infectious Disease 98 (2020) 115078. https://doi.org/10.1016/j.diagmicrobio.2020.115078
- Antibody tests in detecting SARS-CoV-2 infection: a meta-analysis. Kontou, PI et al. Diagnostics 2020, 10, 319; doi:10.3390/diagnostics10050319